Identifier genes in microbial communities
by Suni Mathew
Our lab focuses on finding members of bacterial as well as fungal communities in plants. It is now easier to detect microbes by looking at their genetic sequences rather than identifying microbes morphologically. However, it is laborious, time-consuming and practically impossible to identify each microbial species by reading their whole genomic sequencing. With the advances in next-generation sequencing, microbial genera/species are detected by isolating and monitoring the sequence of an identifier gene.
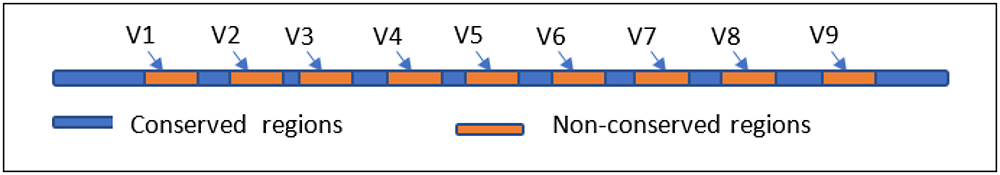
For bacterial community, the identifier gene is the gene coding ribosomal RNA for small subunit 16S widely known as 16SrRNA gene or 16SrDNA. The gene consists of intermittent conserved and variable regions. The gene sequences of conserved regions are same in all bacteria. The nine hypervariable regions named as V1-V9 (Fig.1). The hypervariable regions are unique to each bacterial species, hence by identifying the sequence of the variable regions it is possible to categorize bacterial taxa.
The Fungal community is identified by Internal Transcribed Sequence (ITS) region, present
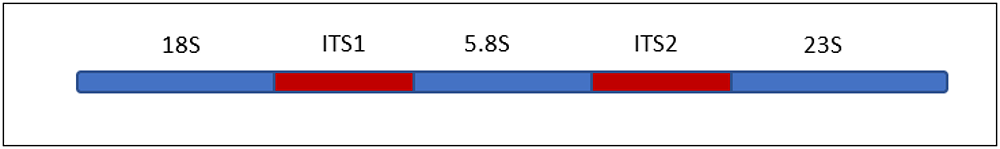
between the ribosomal subunits of eukaryotes (Fig.2), specific for each fungal species.
Once the plant samples are obtained, DNA is extracted using DNA extraction kits and the DNA obtained is a mix of both plant and microbial DNA. Trying to pick 16SrDNA of each bacterial species from this pool of DNA is like searching a needle in a haystack. Thanks to Kary Mullis and his invention of PCR (Polymerase Chain Reaction), that isolation of any gene can be done in a few hours. PCR utilizes chemically synthesized DNA sequences of 10-15 nucleotides called primers as bait to fish out desired genes. The primers for 16SrDNA are designed from the conserved regions. Remember, they are the same for all bacterial species, but the region in between is specific for each bacteria making this interesting. Hence, the same primer acts as a bait to fish out all 16SrDNA in the pool. We will look further into PCR and how to tag 16SrDNA from each microbial taxa.