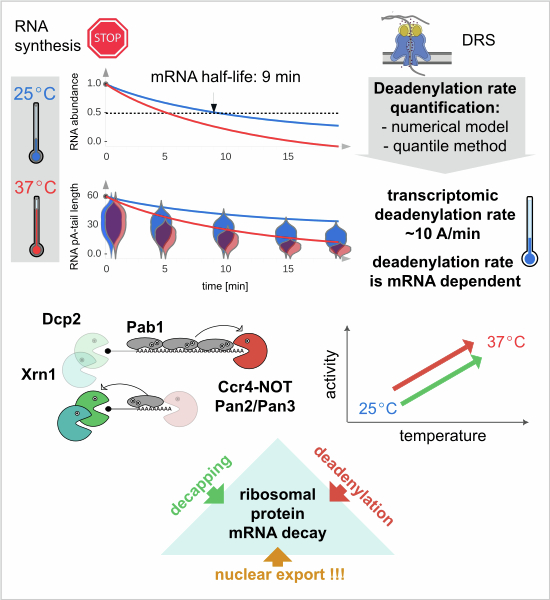
Paper out in EMBO J on modelling mRNA deadenylation rates
We explored the process of mRNA degradation in yeast, focusing on the role of deadenylation (removal of polyA tails) in triggering mRNA decapping and decay. Using direct RNA sequencing and mathematical modeling, we found that deadenylation rates vary across different mRNA types and accelerate during stress. Interestingly, ribosomal protein-coding mRNAs can still be degraded without full deadenylation, relying on active nuclear export, suggesting a more complex relationship between deadenylation and mRNA decay than previously thought. This team effort was led by Agnieszka Tudek who works in Warsaw at the Institute of Biochemistry and Biophysics, Polish Academy of Sciences.
Czarnocka-Cieciura A, Poznański J, Turtola M, Tomecki R, Krawczyk P, Mroczek S, Orzeł W, Saha U, Jensen TH, Dziembowski A, Tudek A (2024) mRNA deadenylation modeling at permissive and stress conditions reveals complex relations to decay. EMBO Journal. https://doi.org/10.1038/s44318-024-00258-3