Research
Processing and assembly of mRNPs
The biogenesis of RNA is a complex process that involves the coordinated activities of transcription machinery (transcription factors and RNA polymerase) and RNA processing factors (such as the capping enzyme, spliceosome, and cleavage and polyadenylation complex). Additionally, a multitude of RNA-binding proteins decorate the RNA, resulting in the formation of ribonucleoprotein particles (RNPs). Of particular interest are messenger RNAs (mRNAs), which serve as genetic information carriers between the nucleus and the cytoplasm. During their synthesis mRNAs assemble into mRNPs, and this process impacts how the mRNA is subsequently processed, transported, translated and degraded. Therefore, understanding the molecular principles that underlie mRNP assembly is critical to unraveling the intricacies of gene expression pathways.
Our research aims to shed light on the mechanisms that govern the coupling of mRNP assembly and RNA processing, and delineate how successful (or unsuccessful) mRNP assembly impacts its transit within the gene expression pathways. Our current studies are focused in the interplay of mRNA 3′-end formation and mRNP assembly. By elucidating these molecular principles, we hope to gain a better understanding of the complex process of RNA biogenesis and its role in regulating gene expression.
 We employ an integrated biochemical approch to examine RNA biogenesis from the molecular scale to the cellular level. To this end, we reconstitute individual RNA processing steps in vitro from purified proteins and RNAs and then utilise a variety of biochemical and biophysical methods to characterise the processes with molecular detail. We extend our structure-function studies to the yeast Saccharomyces cerevisiae to reveal how these mechanisms impact RNA production at the cellular scale.
We employ an integrated biochemical approch to examine RNA biogenesis from the molecular scale to the cellular level. To this end, we reconstitute individual RNA processing steps in vitro from purified proteins and RNAs and then utilise a variety of biochemical and biophysical methods to characterise the processes with molecular detail. We extend our structure-function studies to the yeast Saccharomyces cerevisiae to reveal how these mechanisms impact RNA production at the cellular scale.
Fungal-specific gene expression pathways
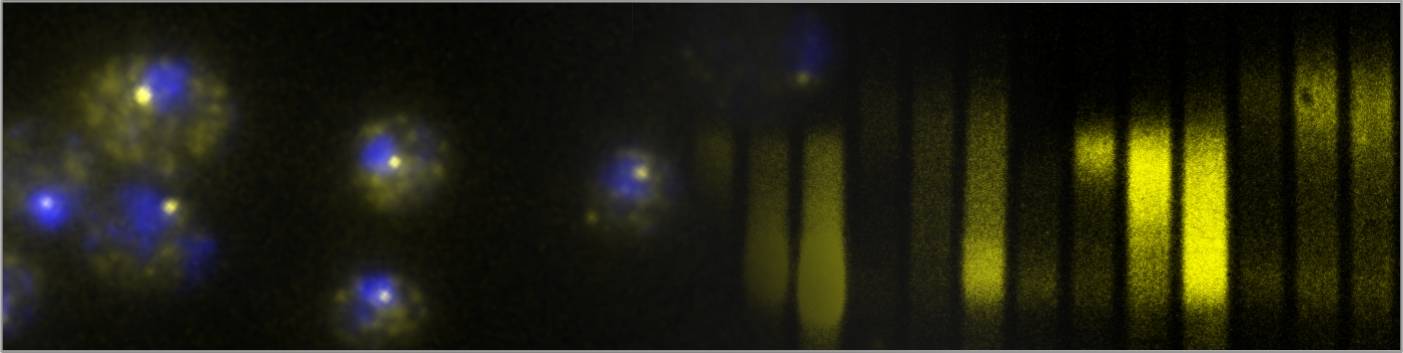
Fungal infections are a significant global health concern, affecting 300 million people and leading to 1.5 million deaths annually. Treatment options are limited, with only three classes of antifungal drugs currently available. The emergence of drug-resistant pathogenic fungal strains further highlights the need for identifying new molecular drug targets. Although many antibacterial compounds target essential steps in bacterial gene expression, identifying fungal-specific targets for chemical inhibition is challenging due to the high level of gene expression machinery conservation across eukaryotes. However, the divergence in mRNA polyadenylation pathways between mammals and fungi (see the figure below) suggests that factors controlling mRNA 3′-end formation could be promising targets for developing urgently needed antifungal therapies. Our research will explore the mechanisms of gene expression regulation to uncover fungal-specific steps in essential processes, with the ultimate goal of identifying new therapeutic targets and developing compounds to inhibit them.

mRNA 3′-end processing pathways differ between yeast (A) and mammals (B). Image from Rodríguez-Molina & Turtola (2023).
Funding