Yeast measure mRNA poly(A) tails with a “kinetic ruler”
The first research paper from our lab uncovers a surprising mechanism for how the yeast Saccharomyces cerevisiae determines the length of messenger RNA (mRNA) tails. Instead of measuring their physical size, cells time their growth with stopwatch-like precision. This “kinetic ruler” reveals a new principle of how molecular accuracy is achieved in essential processes such as gene expression.
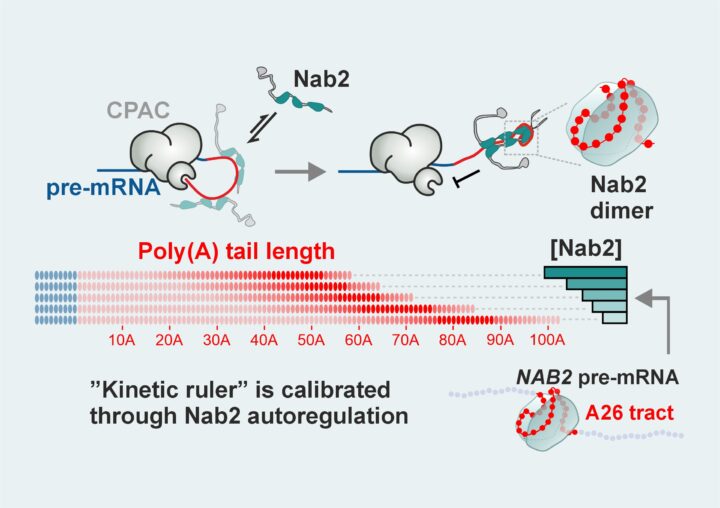
mRNA molecules carry genetic instructions from DNA and end with a poly(A) tail, a protective string of adenosines that controls how much protein is produced and how long the message remains active. In yeast, these tails are consistently about 60 adenosines long, but until now it was unclear how this precision was achieved.
Together with Lori Passmore and Stephen McLaughlin at the MRC Laboratory of Molecular Biology in Cambridge and Torben Heick Jensen inAarhus University, we reconstructed the tail-making process in the laboratory. We found that two players set the length: CPAC, the complex that adds adenosines, and Nab2, a protein that binds to the tail and stops its growth. Once the tail reaches the right length, two Nab2 molecules pair up and block further extension.
However, the final length is set not by the physical dimensions of the tail, but by a race between CPAC’s speed of adding adenosines and Nab2’s speed of binding. The system works like a molecular stopwatch: tail growth always stops between two and three seconds after it begins. Here the concentration of Nab2 in the cell nucleus determines when tail extension stops. Significantly, Nab2 regulates its own levels to keep the timing precisely calibrated even under changing conditions.
This timing-based “kinetic ruler” is critical because the starting length of the poly(A) tail determines how much protein an mRNA can produce and how long the message lasts. By ensuring every new mRNA gets the right-sized tail, cells keep protein production under control.
Similar kinetic rulers may operate in many other biological processes, and understanding them could help explain how disruptions in timing lead to disease. In humans, the Nab2 counterpart ZC3H14 is required for brain development, hinting that these timing-based mechanisms may also be essential for maintaining balanced gene expression in human cells.
Our study was published in Genes & Development (online ahead of print, August 2025).
Link to the article: https://genesdev.cshlp.org/content/early/2025/08/22/gad.352912.125